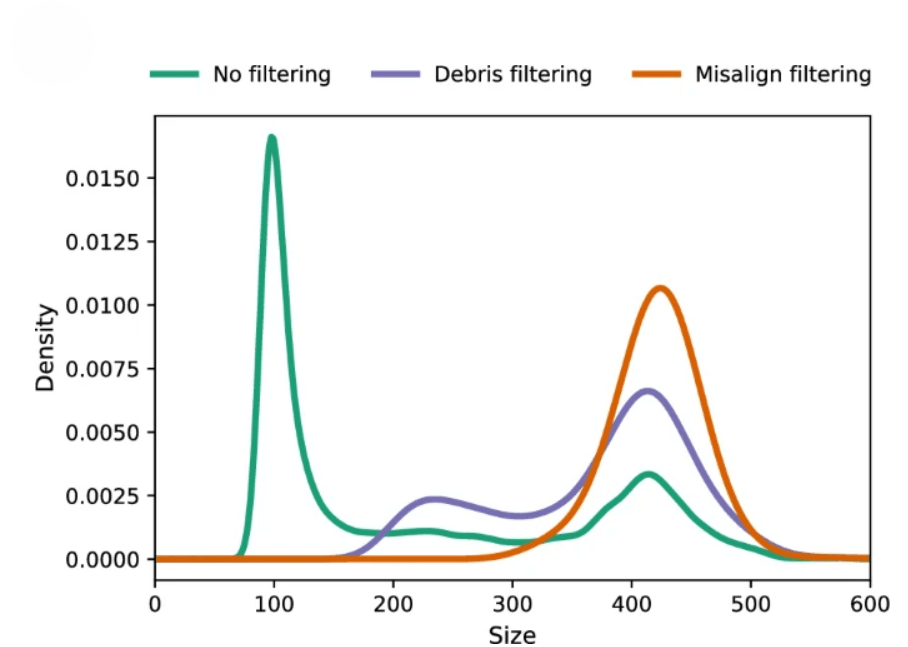
Barghi N and Ramirez-Lanzas C (2023). A high throughput method for egg size measurement in Drosophila. Scientific Reports 13, 3791
Barghi N*, Hermisson J*, Schlötterer C* (2020). Polygenic Adaptation–a unifying framework to understand positive selection. Nature Reviews Genetics 21(12):769–781. *: equal contribution

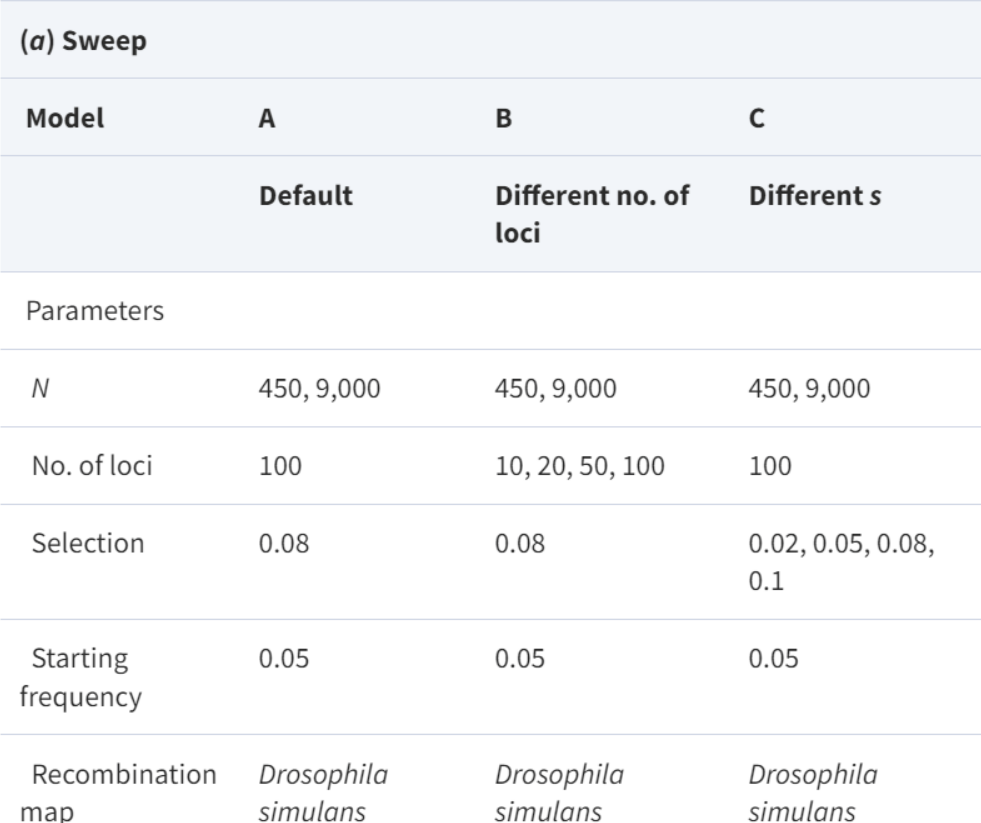
Barghi N and Schlötterer C. (2020) Distinct patterns of selective sweep and polygenic adaptation in evolve and resequence studies. Genome Biology Evolution 12(6):890–904
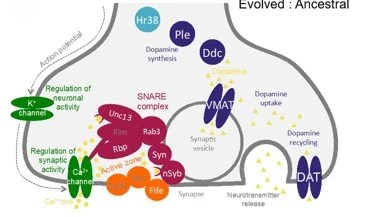
Jakšić AM, Karner J, Nolte V, Hsu S-K,Barghi N, Mallard F, Otte KA, Svečnjak L, Senti KA,Schlötterer C (2020). Neuronal function and dopamine signaling evolve at high temperature in Drosophila. Molecular Biology and Evolution, 37(9):2630–2640
Hsu S, Jakšić AM, Nolte V, Lirakis M, Kofler R, Barghi N, Versace E, Schlötterer C (2020).Rapid sex-specific adaptation to high temperature in Drosophila. eLife 9:e53237
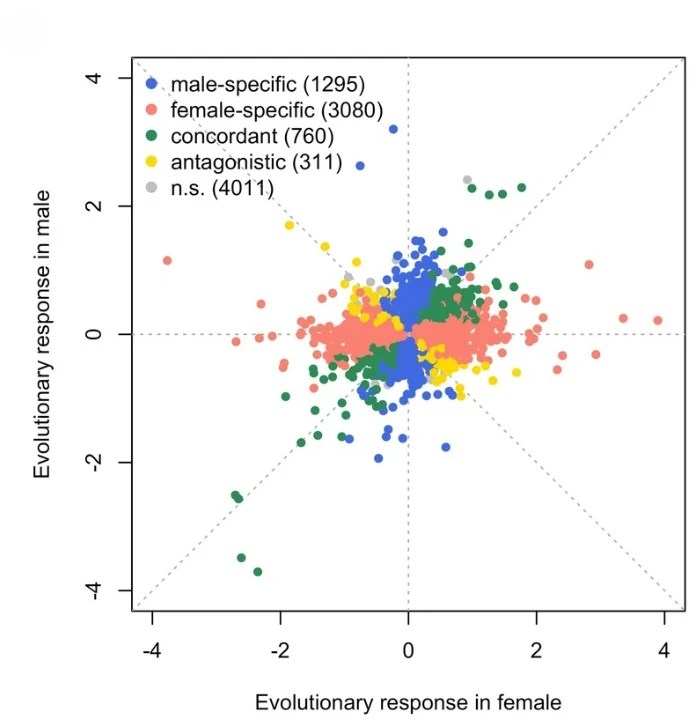
Barghi N, Tobler R, Nolte V, Jakšić AM, Mallard F, Otte KA, Dolezal M, Taus T, Kofler R, Schlötterer C (2019).Genetic redundancy fuels polygenic adaptation in Drosophila, PLOS Biology 17(2), e3000128
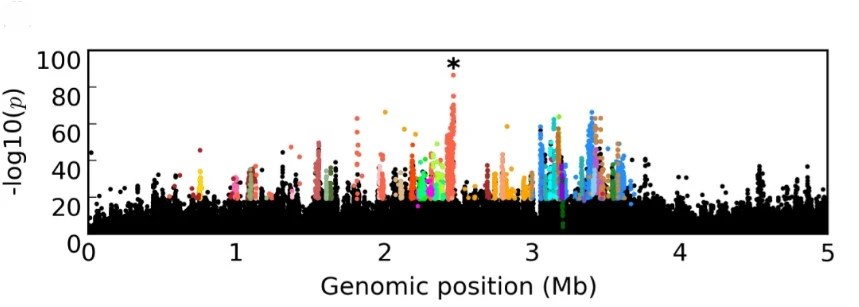
Hsu S, Jakšić AM, Nolte V, Barghi N, Mallard F, Otte KA, Schlötterer C (2019). 24-hour age difference causes twice as much gene expression divergence as 100 generations of adaptation to a novel environment. Genes 10 (89)
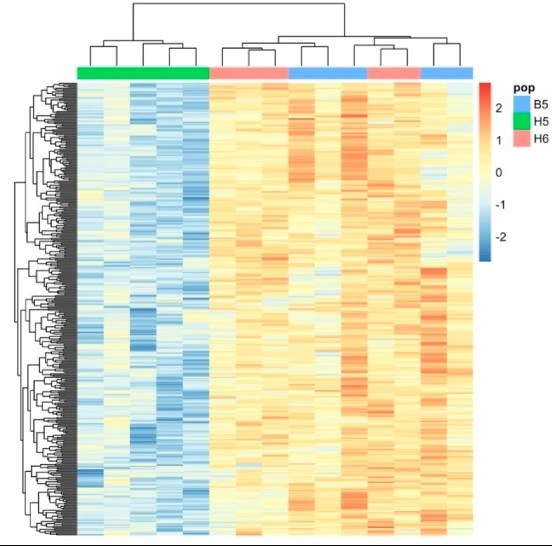
Barghi N, Schlötterer C (2019) Shifting the paradigm in Evolve and Resequence studies: from analysis of single nucleotide polymorphisms to selected haplotype blocks. Molecular Ecology 28(3):521-524

Christodoulaki E*, Barghi N*, Schlötterer C (2019). Distance to trait optimum is a crucial factor determining the genomic signature of polygenic adaptation. bioRxiv, 721340. *: equal contribution.

Nouhaud P, Mallard F, Poupardin R, Barghi N, Schlötterer C (2018) Hight-throughput fecundity measurements in Drosophila. Scientific Reports 8: 4469.
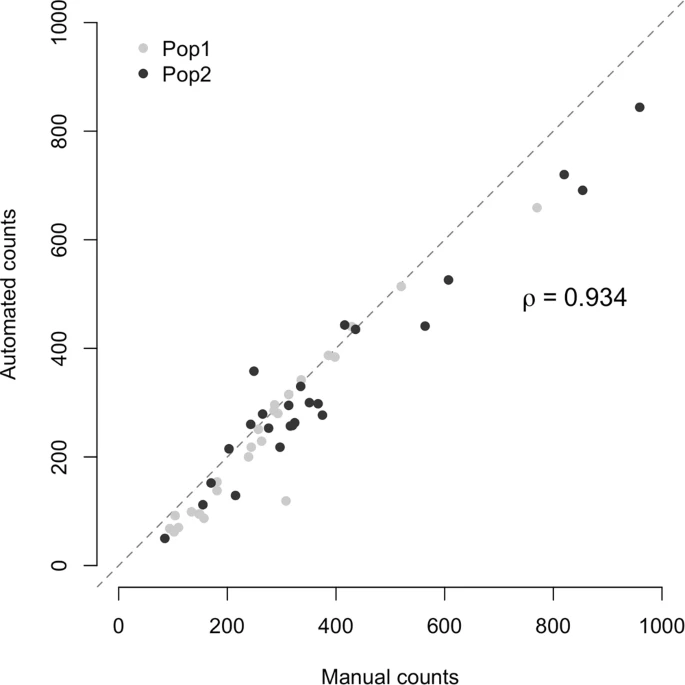
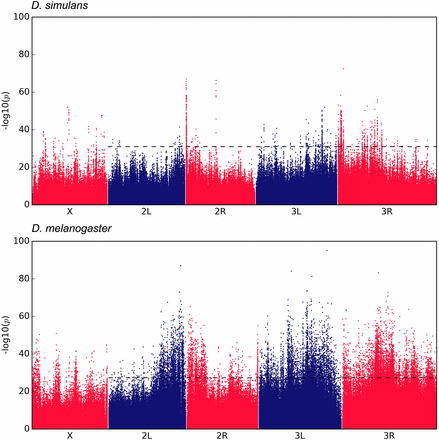
Barghi N, Tobler R, Nolte V, Schlötterer C (2017) Drosophila simulans: A species with improved resolution in evolve and resequence studies. Genes|Genomes|Genetics 7: 2337-2343
Li Q, Barghi N, Lu A, Fedosov AE, Bandyopadhyay PK, Lluisma AO, Concepcion GP, Yandell M, Olivera BM, Safavi-Hemami H (2017) Divergence of the venom exogene repertoire in two sister species of Turriconus. Genome Biology and Evolution 9 (9): 2211–2225.

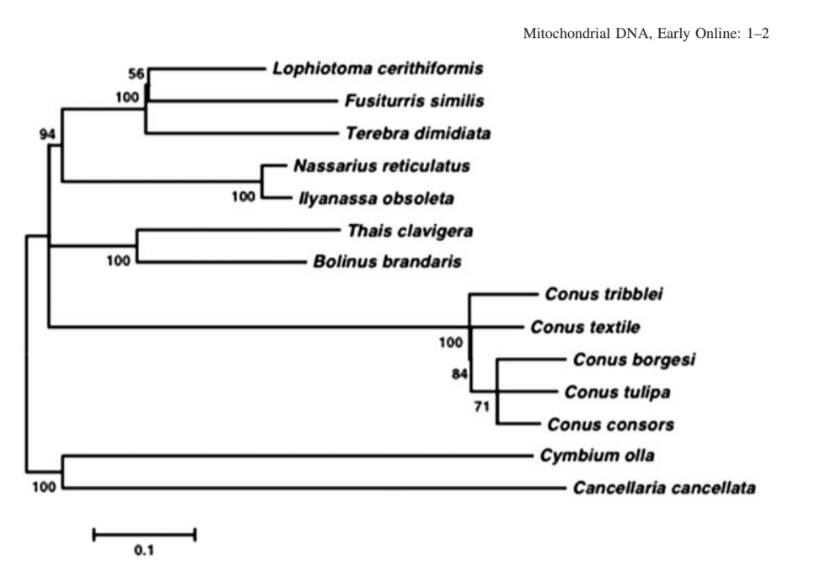
Barghi N, Concepcion GP, Olivera BM, Lluisma AO (2016) Characterization of the complete mitochondrial genome of Conus tribblei Walls, 1977. Mitochondrial DNA 27: 4451–4452
Barghi N, Concepcion GP, Olivera BM, Lluisma AO (2016) Structural features of conopeptide genes inferred from partial sequences of the Conus tribblei genome. Molecular Genetics and Genomics 291(1): 411-422
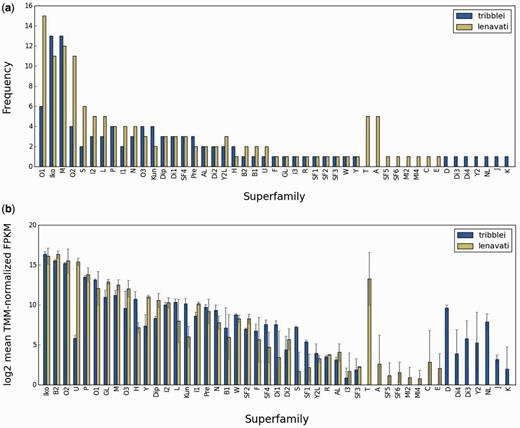
Barghi N, ConcepcionGP, Olivera BM, Lluisma AO (2015) Comparison of the venom peptides and their expression in closely related Conus species: insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biology and Evolution 7 (6):1797-1814

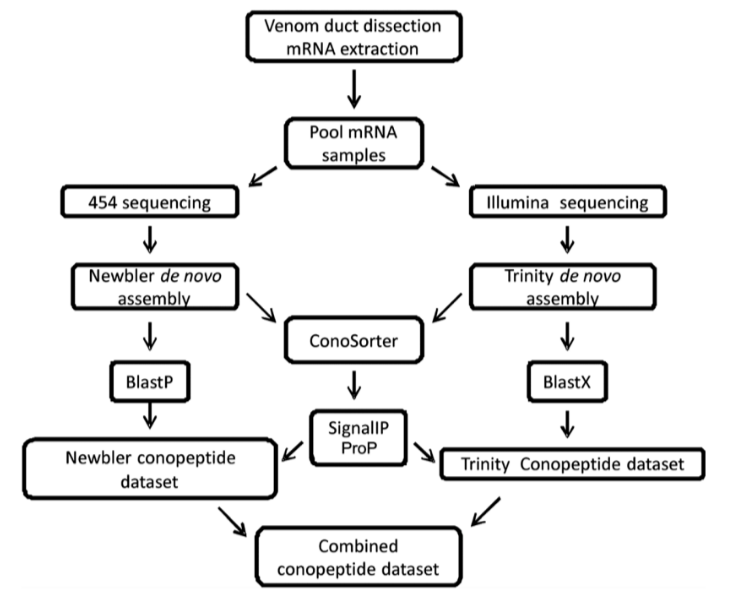
Barghi N, Concepcion GP, Olivera BM, Lluisma AO (2015) High conopeptide diversity in Conus tribblei revealed through analysis of venom duct transcriptome using two high-throughput sequencing platforms. Marine Biotechnology 17 (1): 81-98)